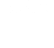Overview
Since its inception, our Scientific Focus Area (SFA) has focused on systems biology of complex bioenergy-relevant microbial communities, ultimately seeking a predictive and quantitative understanding of these systems. Algal ponds and cellulosic bioenergy crops are inherently engineered ecosystems, subject to a range of factors that affect production, many of which are strongly linked to microbial processes. We hypothesize that a better understanding of the constraints on these microbial functions, their genomic underpinning, and their ecological context, will enable scientists and engineers to improve crop stability and predictability, increase efficiency of nutrient use and biomass production, reduce the need for exogenous inputs (for example, fertilizer, carbon dioxide, and water), and design effective strategies to combat pathogens, in order to cultivate sustainable energy crops with predictable and reliable cumulative yields. Our goal is to discover cross-cutting principles that regulate formation and maintenance of phototroph–microbial interactions and their system-level resource allocation consequences, in order to develop a general predictive framework for ecosystem-level impacts of microbial partnerships.
Study systems
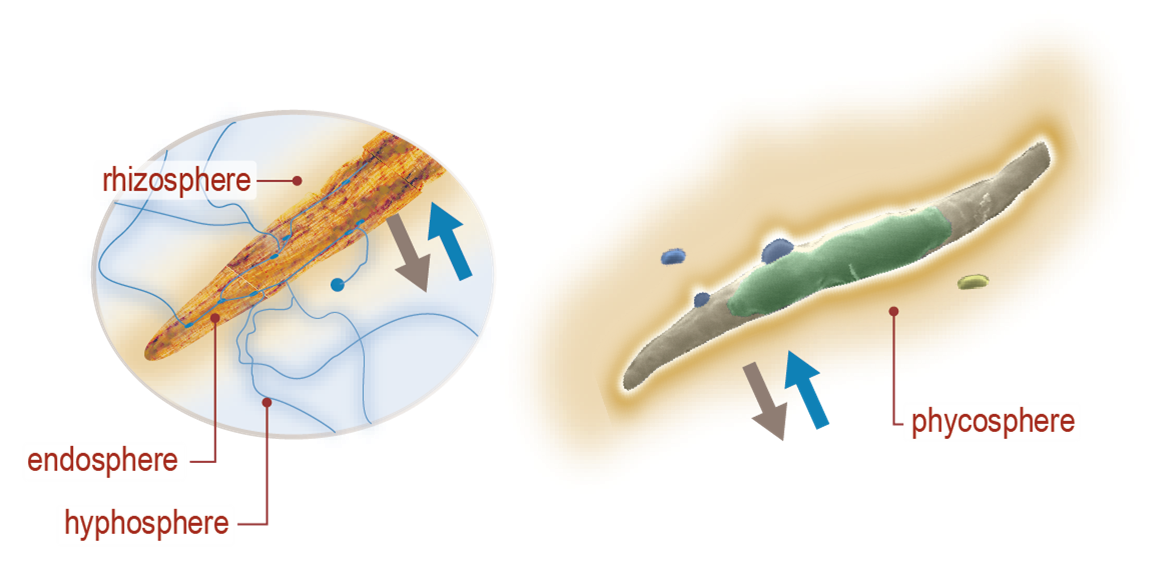
Our current study systems include microalgae and perennial grasses. The µBiospheres SFA is focused on regions of direct interaction between bioenergy-relevant phototrophs and their heterotrophic microbial associates. For algae, this is the phycosphere; the zone on and around individual algal cells, where algae derive most of their nutrients, and where algal exudates that feed surrounding heterotrophs are at their highest concentration. In plants, multiple interaction zones influence plant health and productivity: the rhizosphere zone surrounding roots is a particularly critical nexus of microbial interactions, nutrient exchange, carbon flow, and trophic connections; the hyphosphere zone surrounding fungal hyphae extends the rhizosphere through fungal symbionts into soil; and the endosphere is the more controlled and selective interior environment of plant tissues which mediates nutrient exchange, plant chemistry, and physiology.
Algal–bacterial interactions
Algal–bacterial interactions, ranging from mutualistic to commensal to parasitic, have ecological consequences at all scales and profoundly influence community function and biogeochemical cycling. The variety of algal–bacterial interactions identified in natural systems undoubtedly also occurs in algal biofuel systems, especially open ponds, and the potential to manipulate them for economic benefit is acknowledged. However, the dynamics of pond microbial communities are not well understood, and the consequences of these dynamics on algal productivity are virtually unknown. Our SFA research is designed to produce a mechanistic understanding of specific interactions between individual algal and bacterial strains under controlled laboratory conditions, outdoor mesocosms, and large-volume ponds, significantly improving our understanding of the functional roles of bacterial communities in algal production systems.
Examples of our research in this area include:
- Quantification of carbon and nitrogen exchange between algae and attached bacteria using isotope probing and single cell isotopic analyses (NanoSIMS) in both field and co-cultivation (Arandia et al., 2017, De-Bashan et al., 2016)
- Applying Chip-SIP, a taxon-specific Stable Isotope Probing (SIP) approach, to identify phylogenetic signal in algal organic carbon uptake (Mayali & Weber, 2018)
- Applying NanoSIP (SIP on single cells) on phycosphere enrichment cultures to quantify how bacterial attachment impacts single cell algal carbon fixation rates (Samo et al, 2018)
- Examining community assembly and selection of the attached algal microbiome over time (Kimbrel et al, 2019)
- Light impacts on algal carbon and nitrogen degradation and the importance of bacterial photoheterotrophy in algal microbiomes (Consarnau et al, 2019)
- Bacterial communities can protect against algal grazing (Fisher et al, 2019)
- Algal exometabolomics: Bacterial identity and interaction type impacts exometabolites and diverse algae secrete distinct effector metabolite profiles (Chorazyczewksi et al, 2021; Brisson et al, 2021)
Plant–microbe interactions

Plants are naturally colonized by microbial communities, and these interactions are essential to plant health, soil fertility, and ecosystem function. In bioenergy systems, microbial symbionts can have direct effects on plant productivity, resilience, or stress tolerance of the feedstock crop. However, plant microbial interactions span from mutualistic to parasitic, and the nature of these symbiotic relationships can change depending on current environmental conditions. Our SFA research seeks to understand the fundamental mechanisms underpinning symbioses associated with perennial bioenergy grasses in order to enable sustainable and predictable bioenergy production. We take a systems biology approach that examines multiple scales of interaction, from quantifying microscale nutrient and metabolic exchanges that occur at the interface between plants and microbes, to field scale experimentation that will provide a predictive framework for how symbioses mediate bioenergy sustainability in real-world conditions.
Examples of our research in this area include:
- A comprehensive review of switchgrass microbiomes and their function (Hestrin et al, 2020)
- A soil metatranscriptome study applying the microbial guild concept to show temporal and spatial niche differentiation in the rhizosphere (Nuccio et al, 2019)
- Switchgrass whole-plant microbiome characterizations across natural gradients (Lee et al, 2020)
Research approach
Our SFA uses a “community systems biology: approach, recognizing that the functional properties of most systems are strongly dependent on interactions amongst many taxa. Even highly managed systems, like algal bioenergy ponds or agricultural stands of perennial grasses, are open and classically “complex systems,” and need to be understood from a system perspective. Thus, our work necessarily spans from individual organisms in culture, to constructed consortia, to enrichments, and to complex natural communities.
Isotope tracing
- High-resolution imaging mass spectrometer (Cameca NanoSIMS 50) to perform SIP at the single cell and sub-micron scale, via “nanoSIP.” NanoSIP allows in situ imaging of cell-cell exchange, preserving physical arrangement in a range of complex samples. (Pett-Ridge and Weber, 2012 and Pett-Ridge and Weber, 2021)
- “Density gradient” SIP allows quantitative assessment of substrate uptake and sharing (after incubation with a compound such as 13C-bicarbonate) and can also identify taxon-specific activity (tracing 15NH4+ and/or H218O) by measuring isotope incorporation into DNA or RNA. We built a high-throughput SIP (HT-SIP) pipeline at LLNL that is substantially faster, with 7x less manual effort than the standard approach. HT-SIP can be fully integrated with both metagenomics and quantitative SIP (qSIP), a recent improvement to SIP that estimates taxon-specific growth and death rates for all prokaryotic taxa within natural assemblages.
- Isotope probing using “Chip-SIP,” a custom microarray platform combined with NanoSIMS analysis that can identify taxon-specific isotope incorporation into rRNA. (Mayali et al. 2012).
- Biological accelerator mass spectrometry (BioAMS), which uses a custom moving wire sample introduction system coupled to liquid chromatography separation and AMS, enabling high sample throughput for high sensitivity metabolomics.
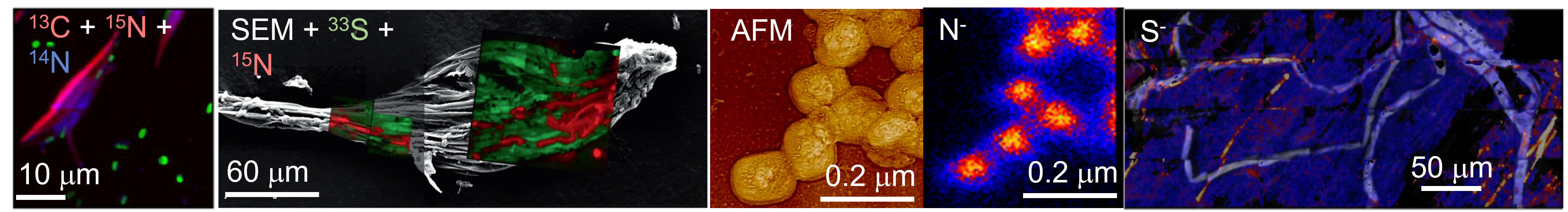
Representative NanoSIP images. Left to right: bacteria associated with P. tricornutum algae, illustrating carbon and nitrogen exchanges; sulfue and nitrogen exchange between moss and cyanobacteria; correlated AFM and nanoSIP depth profile of Vaccinia virions; AMF transport of ammonia to plant root.
Computational biology
Our approach to systems biology also relies on LLNL’s exceptional computing facilities and our in-house informatics and modeling expertise.
Genome Scale metabolic modeling
Genome-scale constraint-based reconstruction and analysis (COBRA) modeling methods (for example, Flux Balance Analysis-FBA) are key tools for complex systems biology analyses and allow us to examine systems at the omics scale. We develop and apply multiple modeling approaches in order to accurately predict metabolic phenomena under variable environmental conditions:
- GX-FBA modeling methodology, which uses gene-expression or proteomic data to constrain FBA models (Navid et al. 2012).
- To model multi-cellular systems and examine the tradeoffs among the competing objectives of the microbial community members, we developed tools for genome-scale multi-objective flux analysis (MOFA) of single cells and simple co-cultures (Navid et al, 2019).
- For analyses of complex communities, we developed a tool for high-dimensional MOFA using LLNL’s high performance computing platforms (Greisemer et al, 2021).
- We simulate the behavior of multi-cellular systems using dynamic FBA (DFBA), an essential tool for analysis of interactions among cells and with their environment.
Improving gene annotation for metabolic modeling
The accuracy of predictions from COBRA models are strongly correlated to the completeness of the genome-based metabolic network reconstruction. We showed that combining genome annotation from multiple annotation tools can provide us with a more complete metabolic network reconstruction (Greisemer, 2018). To improve the accuracy of our network reconstruction for our COBRA models, we have developed a series of apps at KBase that allows for importing and combining annotations from multiple sources to generate draft FBA models, and are currently working on new apps to apply probabilistic modeling to further improve annotation pipelines.
Trait-based modeling
We recognize that while constraint-based methods can be used to simulate metabolism within individual organisms or interactions between multiple cells, they do not scale to natural complexity. Therefore, we are using trait-based models (TBM) as an intermediate approach that preserves key mechanistic properties that determine fitness in dynamic systems. TBM approaches were originally developed in the field of plant ecology but have more recently been applied within microbial ecology at various scales, including global oceans. For our TBMs, we represent the growth and metabolism of microbes using extensions of quota models and dynamic energy budget models.