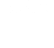September 2023
New publication shows algal exometabolite role in shaping phycosphere bacterial communities
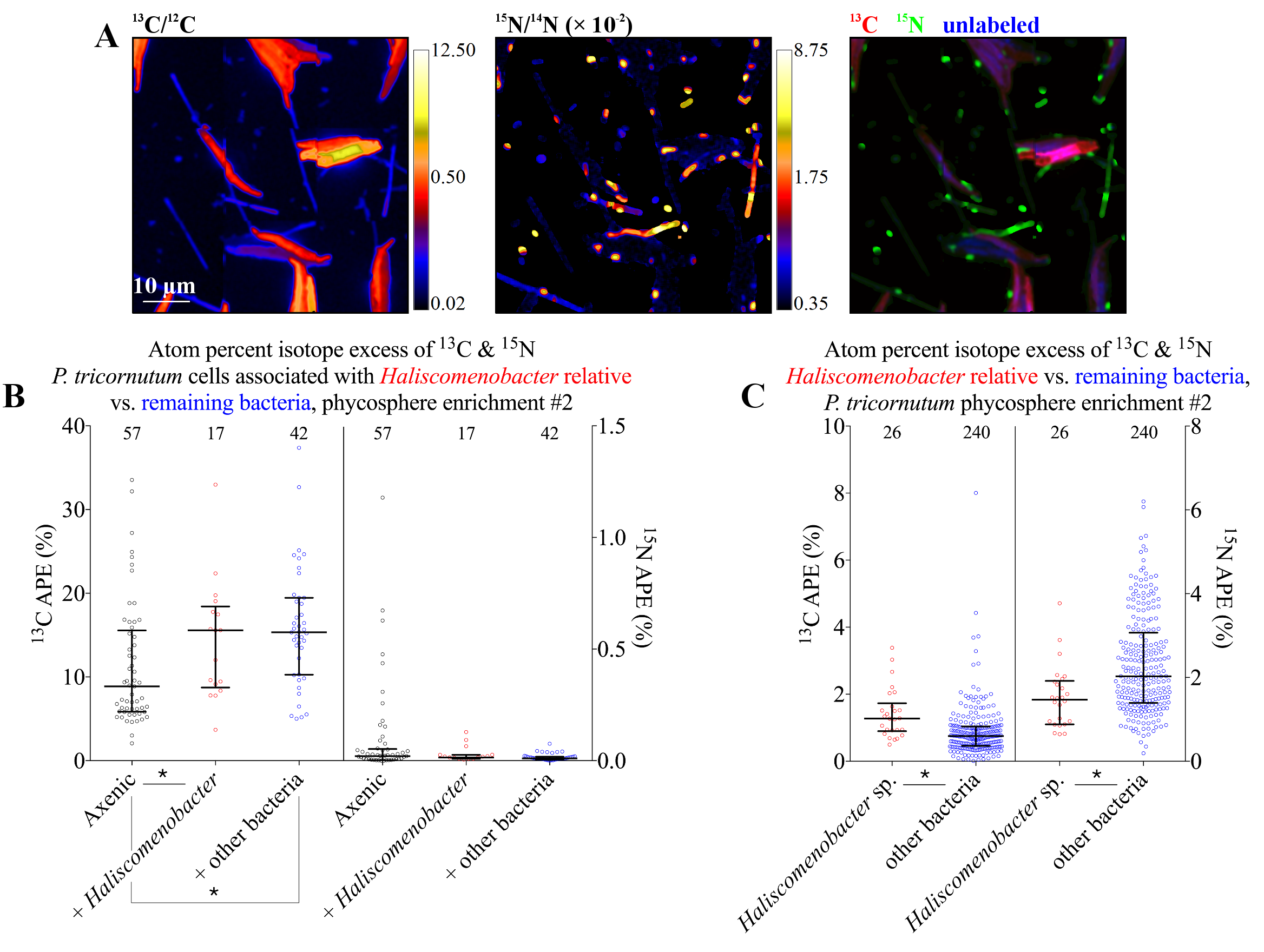
Exometabolites are hypothesized to be a primary driver of host-microbe interactions, but most exometabolites do not have known ecological functions. Exometabolites can be broadly grouped into 3 functional classes: substrate molecules that are resources for microbial biomass production, facilitator molecules that enable chemical reactions, and signaling molecules. In an SFA study recently published in New Phytologist, we used metabolomic profiling to characterize exudates from the diatom Phaeodactylum tricornutum over algal growth and identified a P. tricornutum exometabolite, 4-hydroxybenzoic acid, which can act as a sole carbon source (a substrate molecule) for some phycosphere bacterial isolates. We further showed that this metabolite acts as a selective force on phycosphere bacterial communities, encouraging bacteria that are best able to use that resource. This illustrates the role of diatom substrate exometabolites in actively shaping the phycosphere microbiome through resource allocation.
[V. Brisson, C. Swink, J. Kimbrel, X. Mayali, T. Samo, S.M. Kosina, M. Thelen, T.R. Northen, and R.K. Stuart. Dynamic Phaeodactylum tricornutum exometabolites shape surrounding bacterial communities. New Phytologist 239, 1420 (2023).]
Goodbyes and Hellos!

The SFA said goodbye to two team members: postdoc Megan Morris, who has taken a job with ORISE-ORAU project management, and intern Courtney Swink, who will be starting a PhD program at the University of Exeter Marine Biological Association with Katherine Helliwill studying algal-bacterial interactions. Best of luck Megan and Courtney!
We also welcome new postdocs Di Liang, who will be working on switchgrass-fungal-bacterial interactions, and Naomi Gilbert, who will be working on algal-bacterial-iron interactions, intern Josh White, working on plant-mycorrhizal interactions, and new SGSR Morgan Lindback, who is visiting LLNL through December to work on algal-bacterial-virus interactions.
SFA representation at summer conferences

(top) Rachel Hestrin presenting at MSA. (middle left) Rhona Stuart at MLD7. (middle right) Xavier Mayali at MLD7, signing the rafters of the Scripps Institution of Oceanography Forum, a tradition for former graduate students. (bottom) Rachel Hammer presenting at ESA.
Our team members presented SFA research at numerous conferences this summer, including Gordon Applied and Environmental Microbiology (R. Hestrin and N. Gilbert), Molecular Life of Diatoms (X. Mayali and R. Stuart), Mycological Association of America Annual meeting (J. Pett-Ridge Karling Address, and R. Hestrin), International Conference on the Cell and Molecular Biology of Chlamydomonas (U. Lingappa), Gordon Cell Biology of Metals (R. Boiteau and N. Coffey), Ecological Society of America (R. Hammer), and the JGI Annual User Meeting (V. Brisson).
November 2022
New SFA publication shows detection of a new class of non-photosynthetic nitrogen fixation

SFA team members used LLNL’s NanoSIMS instrument to detect an alternative class of nitrogen-fixing organism which was known to be widespread in sunlit aquatic systems, but where direct evidence of nitrogen fixation was lacking. The team, led by LLNL graduate scholar Katie Harding, collected isotopic images of individual cells. Employing novel SFA-enabled methods development, they were able to locate and quantify activity of these non-photosynthetic nitrogen fixers to particles, showing for the first time that activity of this group appears to be limited to an attached lifestyle. For more details, see the LLNL Newsline article.
[K.J. Harding, K.A. Turk-Kubo, E. Wing Kwan Mak, P.K. Weber, X. Mayali, and J.P. Zehr. Cell-specific measurements show nitrogen fixation by particle-attached putative non-cyanobacterial diazotrophs in the North Pacific Subtropical Gyre. Nat Commun 13, 6979 (2022).]
February 2022
New publication characterizing exometabolites from a diverse suite of microalgae
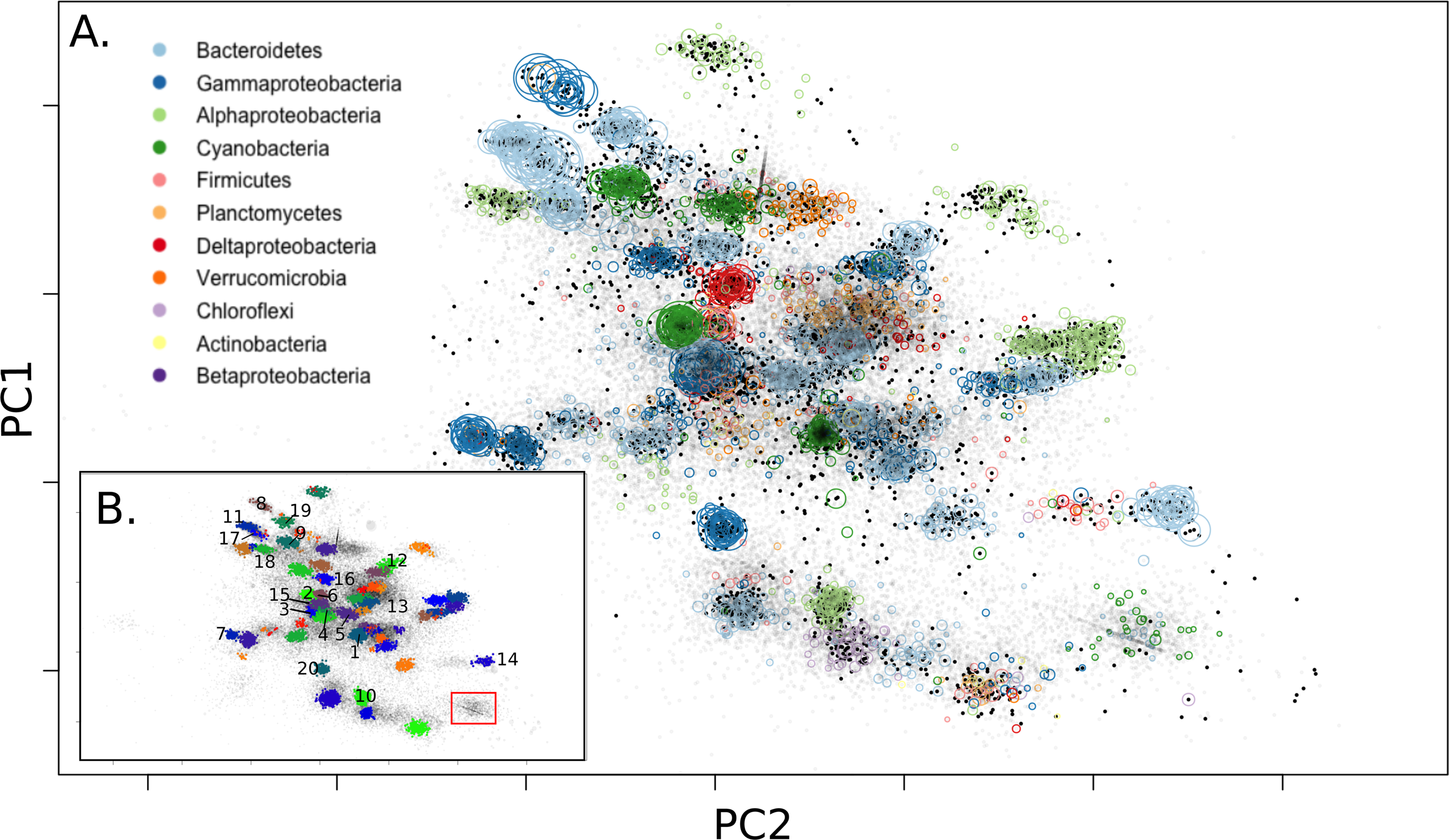
Principal-component analyses of algal exometabolite profiles. Each point represents one of five replicate samples per alga or control condition. Colors indicate which algal culture the sample was from as follows: gray, medium controls; brown, P. tricornutum; light green, M. salina; dark green, C. reinhardtii; light blue, Desmodesmus (saltwater); dark blue, Desmodesmus (freshwater).
Algal exometabolites are important mediators of algal-algal and algal-bacterial interactions that ultimately affect algal growth and physiology. In a recently published manuscript, µBiospheres SFA postdoc Vanessa Brisson led a study which sought to identify effector metabolites that may be involved in interactions within or between algal strains. She profiled exometabolomes across marine and freshwater algae and observed that while phylogeny played a role in exometabolome content, environmental conditions or habitat origin (freshwater versus marine) were also important. She also found that several of these compounds can influence algal growth (as measured by chlorophyll production) when provided exogenously, highlighting the importance of characterization of these novel compounds and their role in microalgal ecophysiology.
[Brisson, V., X. Mayali, B. Bowen, A. Golini, M. Thelen, R.K. Stuart, and T.R. Northen. Identification of Effector Metabolites Using Exometabolite Profiling of Diverse Microalgae. mSystems, 2021. 6(6): p. e00835-21.]
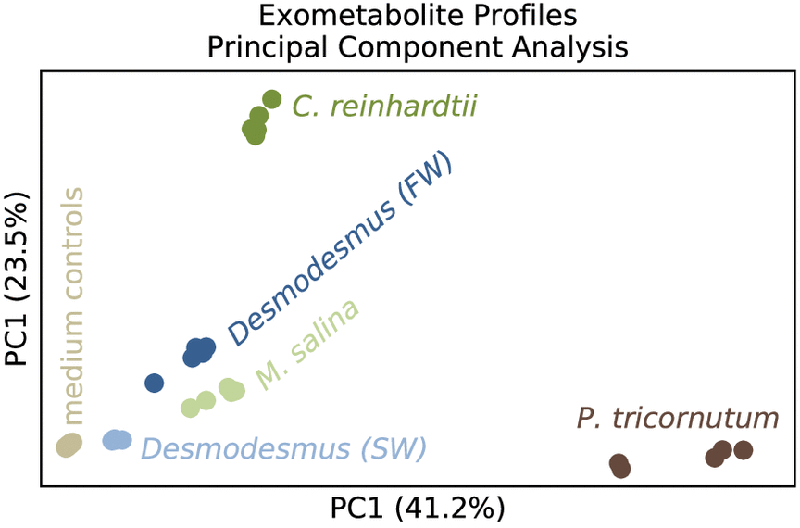
New publication presents a novel co-culture method for algal–bacterial interaction
Schematic diagram of a porous microplate system for monitoring algal–bacterial interaction.
Photosynthetic microalgae and their interaction with microbiomes play a key role in understanding the global carbon cycling and bioenergy production. For decades, it has been hypothesized that the interaction mostly occurs at a local microzone near an alga (“phycosphere”). Aiming to mimic the phycosphere and monitor microbial activities within its region, SFA team members at Lawrence Livermore National Laboratory and the Massachusetts Institute of Technology (co-led by Xavier Mayali, Cullen Buie, and graduate student Hyu Kim) developed a co-culture method—“porous microplate”—that enables the metabolic interaction between algae and bacteria.
With this new protocol, they discovered a model alga Phaeodactylum triconrnutum accumulates to 20 times greater than in a standard culture via a continuous supplementation of nutrients. They also showed that algal-associated bacteria are taxonomically unique in responding to the algal-driven nutrient gradient under the porous microplate. Together, this novel system presents a useful tool to understand a universal microbial interaction in aquatic ecosystems.
[H. Kim, J.A. Kimbrel, C.A. Vaiana et al., Bacterial response to spatial gradients of algal-derived nutrients in a porous microplate, ISME J (2021).]
August 2021
Summer students present research in summer "SLAM" presentations


Summer students Diana Morales (left) and Genevieve Parkey (right).
This summer the SFA hosted two (virtual) summer students, Genevieve Parkey from Georgetown University and Diana Morales (a Mickey Leland Energy Fellow) from Cal Poly Pomona. Both students worked on mathematical models of a Rickettsia-Like Diatom Killer bacterium (RLDK) that can cause algal pond crashes. Genevieve built a genome-scale metabolic model of RLDK from a metagenome-assembled genome (MAG) sequence, and Diana simulated predator-prey dynamics of RLDK in culture with the diatom.
Check out their summer SLAM presentations here:
A view to a kill: System level analysis of metabolism in an algicidal bacterium by Genevieve Parkey
To catch a [diatom] killer by Diana Morales
May 2021
SFA team members presented new annotation tools at KBase webinar

In April, SFA team members Patrik D’haeseleer and Jeff Kimbrel presented at a KBase webinar to showcase the new tools they developed to import, compare, and merge functional annotations for metabolic modeling in KBase. These annotation tools can accept multiple lines of evidence to determine an optimal consensus annotation, allowing annotation data like EC numbers, KEGG, and MetaCyc reaction identifiers to be combined for more accurate draft metabolic models. For more information, watch the webinar Functional Annotation Tools.
Publication shortlisted for The ISME Journal 2020 Best Paper Award

A publication from 2020 led by SFA team members Erin Nuccio and Jennifer Pett-Ridge was shortlisted for the best paper of the year for its “outstanding quality and impact,” and it is included in a special award collection. The publication (described in our June 2020 highlights) focused on using gene expression to identify rhizosphere decomposer guilds, and was one of the first to map microbial gene transcripts to the genomes of individual soil organisms. Congratulations Erin and Jennifer!
SFA team member interviewed on the JGI Podcast Genome Insider
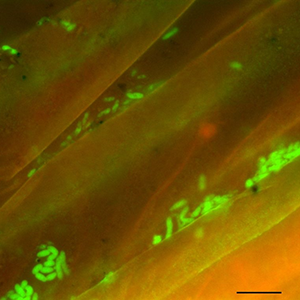
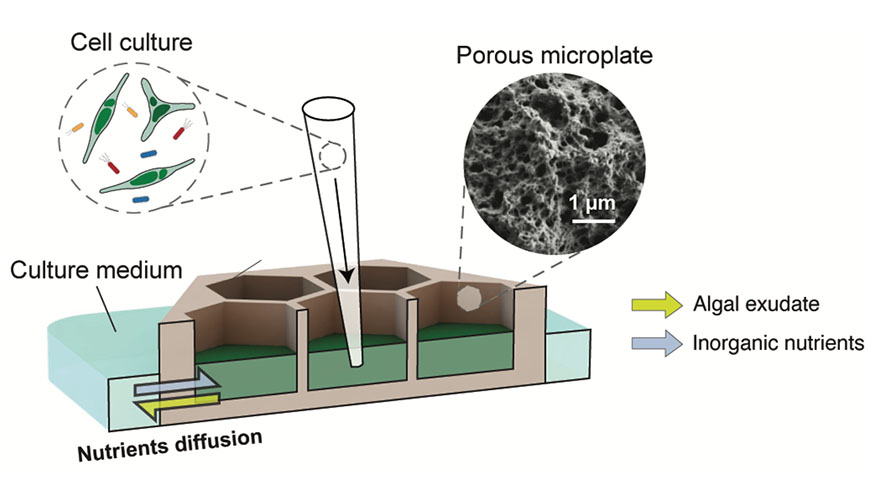
These bright green spots are fluorescently labeled bacteria from soil collected from the surface of plant roots. For reference, the scale bar at bottom right is 10 micrometers long. Photo courtesy of Rhona Stuart (LLNL).
In a recent episode of the JGI podcast Genome Insider, SFA team member Jennifer Pett-Ridge discussed DOE BER Genomic Sciences funded research, including a publication from this SFA examining gene expression to identify rhizosphere decomposer guilds. Read a transcript or listen to the episode entitled “Party in the Rhizoshere.”
February 2021
BioSFA Postdoc Megan Morris wins best paper award from FEMS Microbiology Ecology
Megan Morris (NACS) and co-authors received a 2020 best article award from FEMS Microbiology Ecology for their paper titled, “Microbial abundance, composition, and function in nectar are shaped by flower visitor identity.” The award recognizes the best articles submitted to each of the seven journals founded by the Federation of European Microbiological Societies (FEMS). The FEMS Microbiology Ecology editorial board recognized the paper as “outstanding in terms of ecological question, new concepts, and examining intraspecific diversity and functions.” In addition to a cash prize, the award includes special marketing to promote the authors’ research and an interview with the authors.
September 2020
Virtual summer student presents on her SFA research


LLNL SFA team member Ali Navid hosted a virtual summer student this year from Georgetown University, Samantha Shen. Samantha helped build and curate a genome-scale flux balance analysis (FBA) model for bacterial strain Kordia algicida OT-1. She presented her work at LLNL’s summer student virtual symposium. The model will be used to examine the unique metabolic needs of K. algicida as well as the robustness of its metabolism to genetic and environmental perturbations. We will also examine the benefits, as well as the metabolic burden associated with continual production of algicidal proteins.
New publication: NanoSIMS review
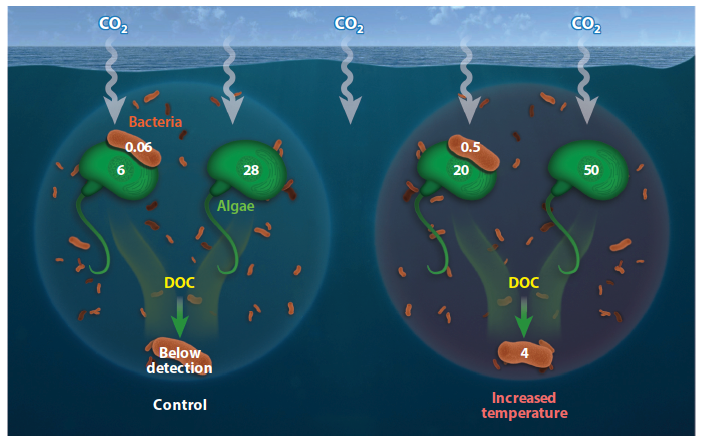
NanoSIMS single-cell isotope incorporation data scaled to the ecosystem level, derived from a study that incubated a marine community at ambient and increased temperatures and measured inorganic 13C incorporation in free-living and attached cells (Arandia-Gorostidi et al. 2017). Unitless numbers represent the total isotope incorporation calculated for heterotrophic bacteria (orange rods) and autotrophic algae (green flagellated cells). Abbreviations: DOC, dissolved organic carbon; NanoSIMS, nanoscale secondary ion mass spectrometry.
LLNL SFA team member Xavier Mayali recently published an invited review in the Annual Review of Marine Science series. This publication presents an update of NanoSIMS-enabled research in aquatic environments including water column and sediments, summarizing the literature from the last few years that focus on both rare metabolism and ubiquitous microbial activity. In addition to highlighting a few novel applications as well as general caveats of NanoSIMS, it attempts to briefly identify new directions for research in the next few decades, including linking NanoSIMS data at the small scale with larger scale ecosystem modeling.
[Mayali, X., NanoSIMS: Microscale quantification of biogeochemical activity with large-scale impacts. Annual Review of Marine Science, 2020. 12(1)]
June 2020
New publication: At the root of the matter: using gene expression to identify rhizosphere decomposer guilds

Rhizosphere microbes have specialized decomposition strategies in distinct root habitats over time and space.
The soil surrounding roots, or rhizosphere, is a critical zone for carbon transformations in the terrestrial biosphere. Stimulated by exudates and root decay, rhizosphere organisms interact to break down carbon derived from root tissues and move it into the surrounding soil, ultimately regulating how soil carbon is stabilized or lost from the system. However, the mechanisms that underpin soil carbon cycling in the rhizosphere are poorly understood.
In a recent publication, SFA team members Erin Nuccio and Jennifer Pett-Ridge used microbial gene expression to determine how roots affect decomposition and identified distinct groups of microorganisms that undertake decomposition during a temporal succession—a process known as niche differentiation. Until now, niche differentiation has been difficult to describe for soil microorganisms due to their high degree of functional redundancy and the fact that most soil microbes cannot be cultured in the laboratory. Partnering with colleagues from UC Berkeley, Lawrence Berkeley National Laboratory, the Joint Genome Institute, and the University of Oklahoma, the group used metatranscriptomics to measure and describe the genes expressed in several soil habitats. The group’s analysis is one of the very first to map microbial gene transcripts to the genomes of individual soil organisms—allowing the authors to determine which specific taxa were inhabiting particular roles or “niches” in the soil, and how these patterns change with time. The results show that organisms primarily belong to three guilds, each of which has different capabilities and metabolic preferences. This work provides a key step towards developing microbially-constrained models to predict the fate of soil carbon.
[E.E. Nuccio, E. Starr, U. Karaoz, E.L. Brodie, J. Zhou, S.G. Tringe, R.R. Malmstrom, T. Woyke, J.F. Banfield, M.K. Firestone, and J. Pett-Ridge, Niche differentiation is spatially and temporally regulated in the rhizosphere, The ISME Journal 14, 999–1014 (2020), doi: 10.1038/s41396-019-0582-x.]
April 2020
SFA team members present at DOE BER’s Genomic Sciences Program Annual PI meeting
At this year’s Department of Energy (DOE) Biological and Environmental Research (BER) meeting, held in February, SFA team members Peter Weber, Rhona Stuart, Patrik D’haeseleer, and Ali Navid presented on data from our SFA during the main session and the follow-on All-SFA half-day meeting.
Peter gave a talk in the Emerging Technologies session entitled “New Insights into Microbial Function and Interactions Using Stable Isotope Probing and NanoSIMS.” Peter showed the breadth of BER research performed under our SFA, the LLNL Soil Microbiome SFA, and related projects using the LLNL NanoSIMS. He highlighted research under our SFA on plant-fungi symbiosis, algal-bacterial interactions, algal metal metabolism, and boreal moss-cyanobacteria symbiosis.
Rhona’s talk entitled “Symbioses in Bioenergy-Relevant Crops; Perspectives from the Single Cell to the System Scale” was in the Systems Biology of Bioenergy-Relevant Microbes Awards session. Rhona presented on algal-bacterial interactions across scales, from complex algal ponds to the co-cultures. Her presentation highlighted the role of algal physiology in influencing the algal microbiome, and a deeper dive into a single algal-bacterial mutualism using single cell isotope tracing and proteomics.
Patrik gave both a talk (“Tools for Importing, Comparing and Merging Functional Annotations for Improved Metabolic Modeling in KBase”) and a hands-on demo on our KBase app development project, illustrating how to import enzyme annotations from DOE JGI’s IMG platform or dozens other 3rd party annotation tools to KBase.
Our SFA also participated in the All-SFA follow-on half-day “Multi-Omics to Models” workshop, where Ali Navid gave a talk titled “Examining phototroph-bacterial interactions by using omics-based computational models.” Ali presented the latest results from our in silico analysis of metabolic changes in an algal-bacterial system using our GX-FBA method for integrating proteomic data into genome-scale models. He highlighted how the results of these studies have led to design of new experiments and identification of a class of compounds that could be responsible for the observed algal phenotypes.
New publication shows the power of nanoSIP to characterize microbial metabolism
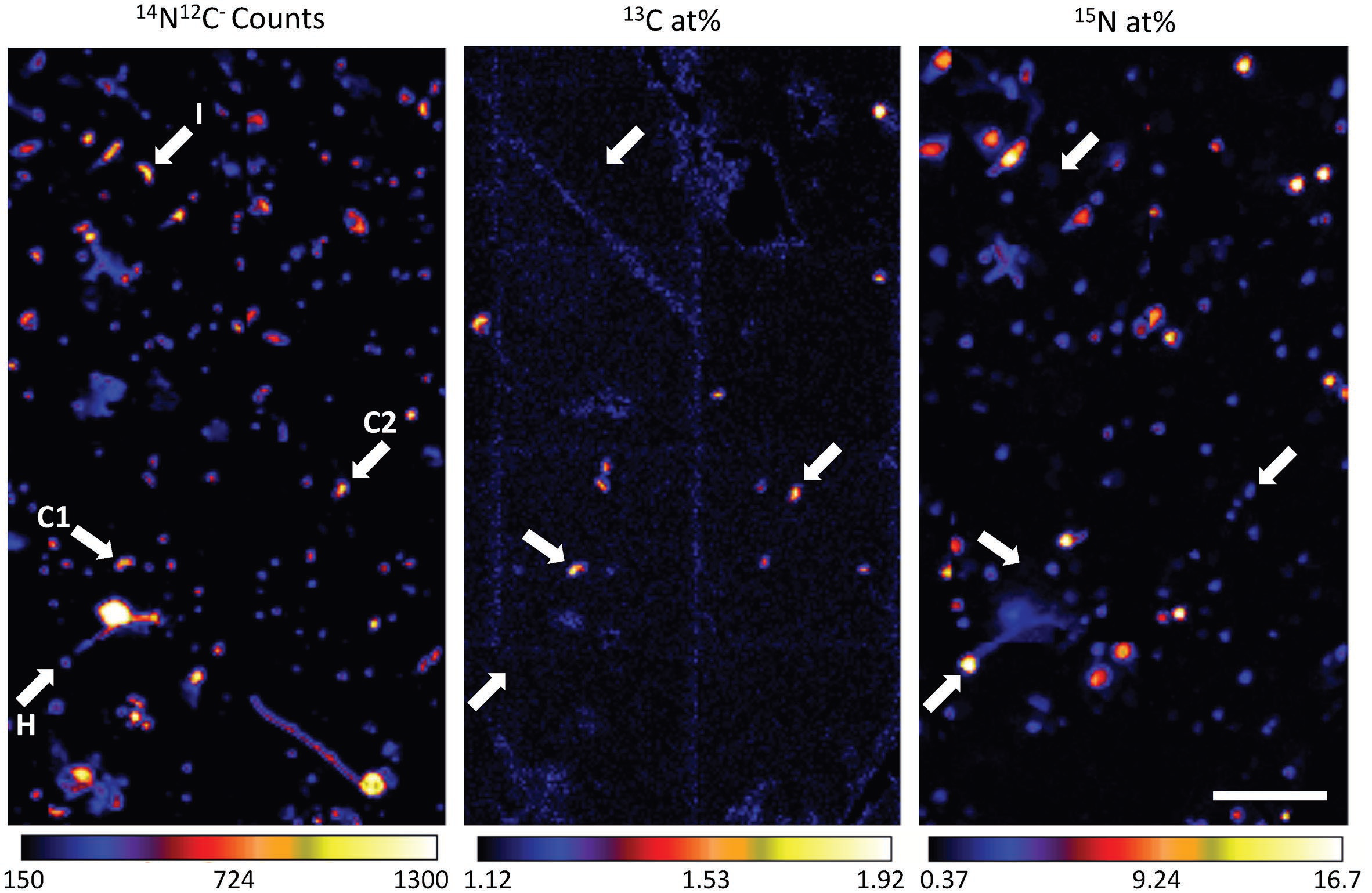
Representative nanoSIP images demonstrating high-throughput metabolic screening of cells filtered from Pacifica, California seawater incubated with 13C-bicarbonate and 15N-amino acids for 6 days. 14N12C- ion counts reflect all carbon- and nitrogen-containing particles, 13C atom percent indicates cells enriched in 13C, and 15N atom percent indicates cells enriched in 15N. The same four cells are indicated with arrows in each panel, with letters in the first panel indicating putative metabolism: I (no enrichment; inactive cell), C1 (enrichment in only 13C; chemoautotroph), H (enrichment in only 15N; heterotroph), and C2, (enrichment in 13C, minor enrichment in 15N; chemoautotroph). Scale bar is 11 μm.
SFA team members Xavier Mayali, Jessica Wollard, Peter Weber and Jennifer Pett-Ridge joined with researchers from Stanford University and the University of Southern California to use multi-isotope nanoSIMS analysis (nanoSIP) to study the anabolic activity of marine microorganisms with an emphasis on natural populations of Thaumarchaeota. They used fluorescence in situ hybridization (FISH) with nanoSIP to confirm that most Thaumarchaeota were living chemoautotrophically, while bacteria were not. FISH-nanoSIP analysis of cells incubated with dual-labeled (13C,15N) amino acids revealed that most Thaumarchaeota cells assimilated amino-acid-derived nitrogen but not carbon, while bacteria assimilated both. This indicates that some Thaumarchaeota do not assimilate intact amino acids, suggesting intra-phylum heterogeneity in organic carbon utilization, and potentially their use of amino acids for nitrification. Together, their results demonstrate the utility of nanoSIP for high-throughput metabolic screening and shed light on the activity and metabolism of uncultured marine archaea and bacteria.
A.E. Dekas, A.E. Parada, X. Mayali, J.A. Fuhrman, J. Wollard, P.K. Weber and J. Pett-Ridge (2019) Characterizing chemoautotrophy and heterotrophy in marine archaea and bacteria with single-cell multi-isotope nanoSIP. Frontiers in Microbiology, doi: 10.3389/fmicb.2019.02682.
January 2020
SFA team member chairs session on harmful algal blooms at AGU Fall Meeting

Yiwei Cheng from Lawrence Berkeley National Laboratory organized and chaired a session at the American Geophysical Union’s Fall Meeting in San Francisco, CA. The session, titled “Harmful algal blooms across river-sea continuum: Understanding its cascading impacts from food webs, biogeochemical cycles to human infrastructure,” was on December 9–10, 2019. Speakers presented relevant work that integrate physical, molecular, chemical, biological, geological observations/experiments, and modeling that advance understanding of HABs biodiversity, species adaptation and interactions, toxin biosynthesis, population dynamics, HABs observation, prediction and development of mitigation strategies.
December 2019
New publication on Volatile Organic Compound biomarkers of algal pond crashes
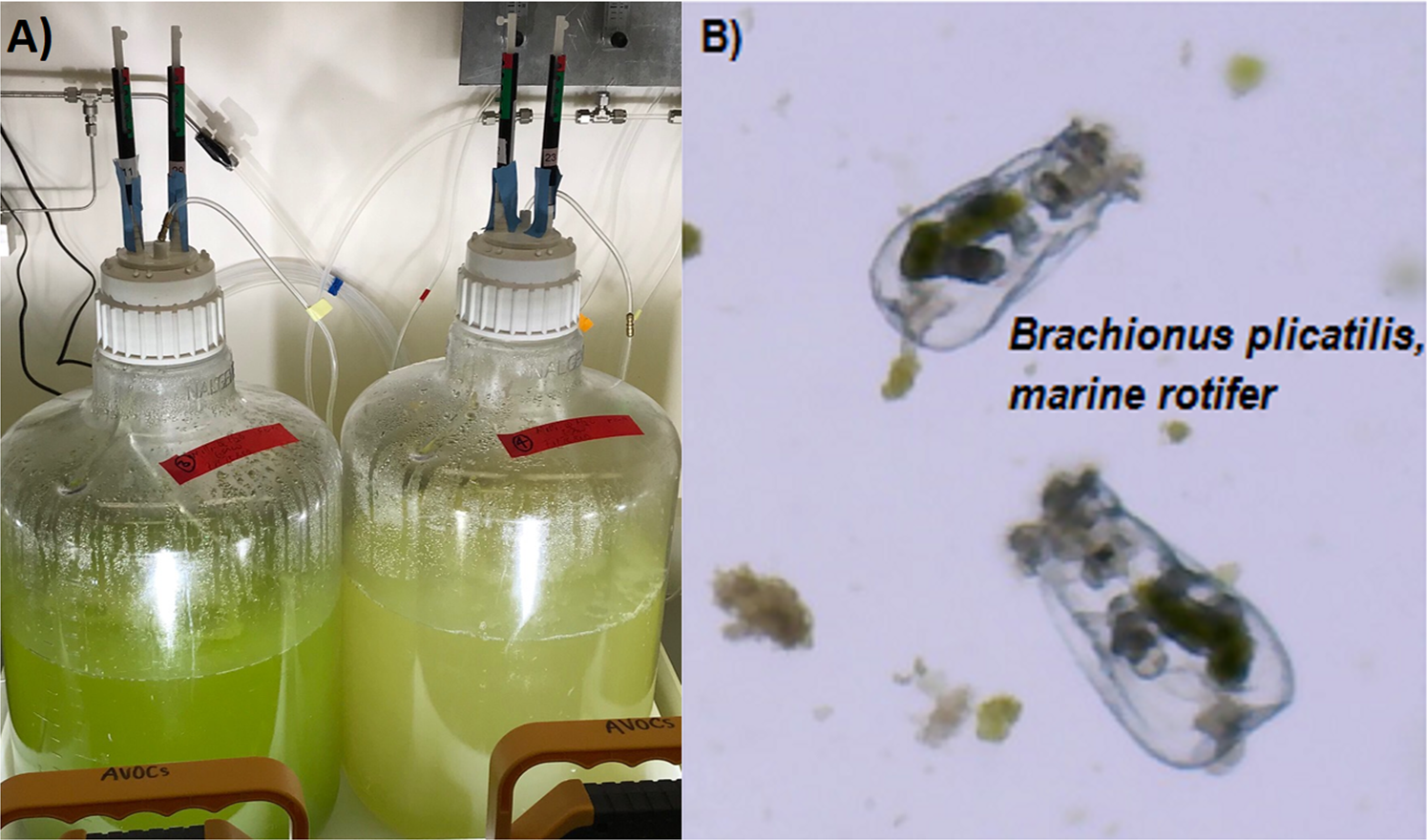
15 L algal mass cultures (analogous to production ponds) with a healthy algal culture on the left compared with a crashed algal culture on the right. (B) Brachionus plicatilis (average length 160 µm), marine rotifer, in a field of microalgae, Microchloropsis salina.
SFA team members at LLNL and Sandia National Laboratory recently published a manuscript on chemical profiling of volatile organic compounds (VOCs) from algal cultures. Using a specially designed algal culturing system to minimize background VOCs and solid-phase microextraction (SPME) sampling coupled with GC-MS, the team conducted an untargeted analysis of VOCs. They compared headspace samples from healthy and rotifer-infested cultures of marine algae Microchloropsis salina, and found distinct differences that indicate specific VOCs released by infected algal cultures. Example VOCs included trans-beta-ionone and beta-cyclocitral, attributed to carotenoid degradation, which were detected in the infected cultures. The VOCs could be used as early indicators for impending pond crashes.
For more information, see the feature in Biofuels, Bioproducts and Biorefining.
[Reese, K., C. Fisher, P. Lane, J. Jaryenneh, M. Moorman, D. Jones, M. Frank, and T. Lane. Chemical Profiling of Volatile Organic Compounds in the Headspace of Algal Cultures as Early Biomarkers of Algal Pond Crashes. Sci Rep 9, 13866 (2019)]
October 2019
New Publication on Mn imaging and metabolism in model alga Chlamydomonas reinhardtii
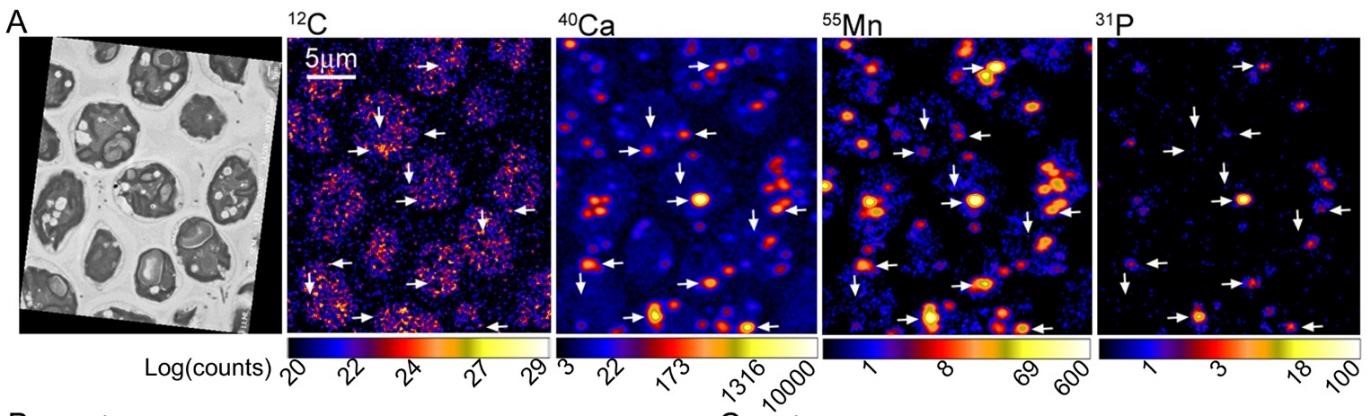
Correlated STEM and nanoSIMS imaging of cells grown in medium containing 1000µM Mn, shows Mn co-localization with P and Ca under Mn hyperaccumulation conditions.
SFA team members contributed to an investigation led by Sabeeha Merchant’s group on Mn metabolism in Chlamydomonas reinhardtii recently published in J. Biol. Chem. It describes the controls on Mn accumulation and identifies polyphosphates as the critical Mn transport mechanism. The team used the LLNL NanoSIMS to map the location of Mn in cells and were able to show its correlation with Ca and P over two orders of magnitude of concentration. The NanoSIMS data are consistent with acidocalcisomes storage.
Tsednee M, Castruita M, Salomé PA, Sharma A, Lewis BE, Schmollinger SR et al (2019). Manganese co-localizes with calcium and phosphorus in Chlamydomonas acidocalcisomes and is mobilized in Mn-deficient conditions. Journal of Biological Chemistry: jbc. RA119. 009130.
New Book Chapter on ChipSIP method published

SFA team members recently published a book chapter on the SFA-developed method Chip-SIP. Chip-SIP is a stable isotope probing (SIP) method for linking microbial identity and function in mixed communities and is capable of analyzing multiple isotopes (13C, 15N, and 18O) simultaneously. This method uses a high-density microarray to separate taxon-specific 16S (or 18S) rRNA genes and a high sensitivity magnetic sector secondary ion mass spectrometer (SIMS) to determine the relative isotope incorporation of the rRNA at each probe location. Using a maskless array synthesizer (MAS), we synthesize multiple unique sequences to target hundreds of taxa at the ribosomal operational taxonomic unit (OTU) level on an array surface, and then analyze it with a NanoSIMS 50, using its high-spatial resolution imaging capability to generate isotope ratios for individual probes. The Chip-SIP method has been used in diverse systems, including surface marine and estuarine water, rhizosphere, and peat soils, to quantify taxon-specific relative incorporation of different substrates in complex microbial communities. Depending on the hypothesis and experimental design, Chip-SIP allows the user to compare the same community incorporating different substrates, different communities incorporating the same substrate(s), or quantify how a community responds to treatment effects, such as temperature or nutrient concentrations.
Mayali X, Weber PK, Nuccio E, Lietard J, Somoza M, Blazewicz SJ et al (2019). Chip-SIP: Stable Isotope Probing Analyzed with rRNA-Targeted Microarrays and NanoSIMS. In: Dumont MG, Hernández García M (eds). Stable Isotope Probing: Methods and Protocols. Springer New York: New York, NY. pp 71-87.
New SFA-synergistic publication on imaging surface Fe distribution from dust in a globally relevant nitrogen-fixing microbe
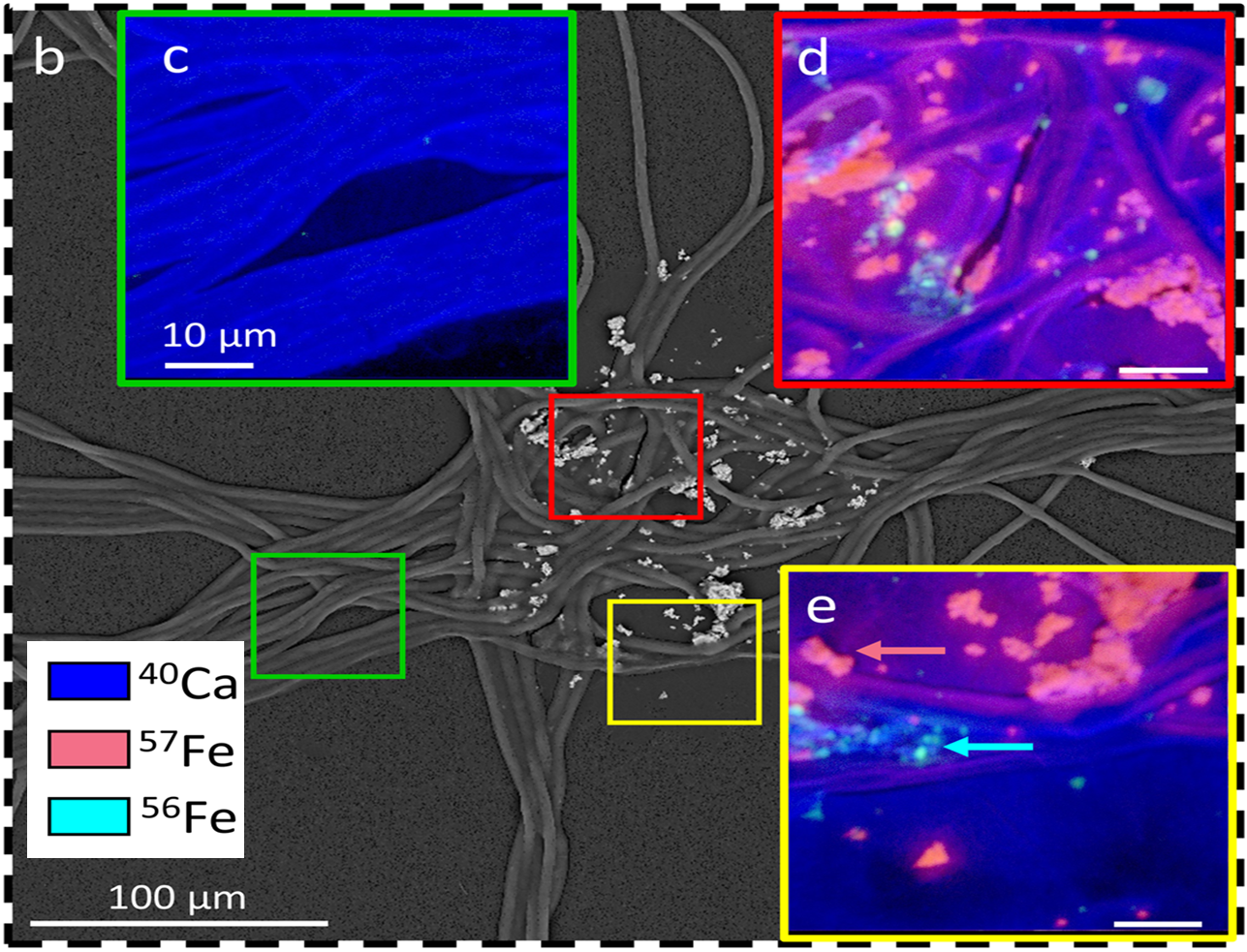
NanoSIMS mapping of 40Ca, 57Fe and 56Fe distributions in a natural Trichodesmium colony incubated with 57Fe-enriched particles, showing centering of newly added 57Fe and retention of older 56Fe particles.
As part of a synergistic LDRD-funded project, SFA team members R. Stuart and P. Weber contributed to a recent publication relevant to our SFA trace metal imaging methods development efforts. The paper found that a globally important nitrogen-fixer, Trichodesmium, is able to selectively collect Fe-rich particles, over other particles, to maximize its Fe supply. To examine this process at the submicron scale, natural Trichodesmium colonies were collected and incubated for 24h with 57Fe-enriched particles to examine particle size distribution and retention within the colony. Using NanoSIMS we mapped surface Fe distribution on the colonies and found a concentration of Fe-rich particles in the center of the colony, which included both the newly added 57Fe and older 56Fe retained prior to collection, indicating centering of these smaller particles sizes.
Kessler N, Armoza-Zvuloni R, Wang S, Basu S, Weber PK, Stuart RK et al (2019). Selective collection of iron-rich dust particles by natural Trichodesmium colonies. The ISME Journal.
August 2019
SFA Lawrence Scholar graduate student wins best poster award

Katie Harding, a University of California at Santa Cruz graduate student conducting research at LLNL with SFA team members, won the best poster award at LLNL’s summer poster symposium for summer interns and continuing student researchers working in many different research areas. Her poster was entitled “Symbiotic unicellular cyanobacteria fix nitrogen in the arctic ocean,” and describes nanoSIP FISH-SIMS research investigating a symbiotic nitrogen-fixing association previously thought to be constrained to subtropical climate. The work shows that these symbiotic microbes fix nitrogen in climate-sensitive Arctic waters as well. Congratulations Katie!
SFA team member presents at 2019 Gordon Applied Environmental Microbiology Conference

SFA team member Ty Samo presented his SFA-funded research at the Applied Environmental Microbiology GRC in South Hadley, MA this July. His poster entitled “Insights on algal-bacterial symbioses mediated by metabolite exchange and organic matter remineralization,” showcased his work on Phaeodactylum tricornutum interactions with a phycosphere-associated bacterium, Algoriphagus sp. ARW1R1, which enhances algal growth. The work shows that one aspect of enhanced growth in P. tricornutum mediated by Algoriphagus sp. ARW1R1 may be generated in part by a positive feedback between microalgal DOM production, bacterial DOM remineralization and niacin production, and stimulation of central carbon metabolism in both partners through increased NAD+ concentrations synthesized from niacin precursors.
New Publication utilizing Chip-SIP to examine the role of photoheterotrophy
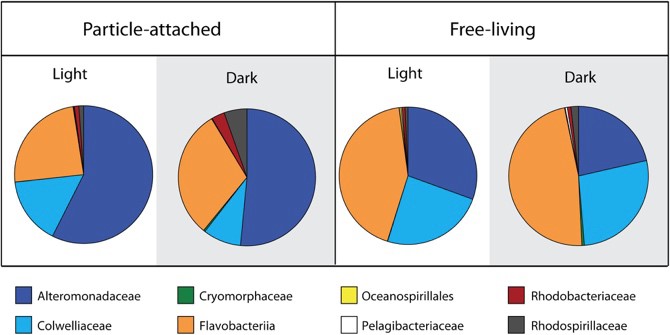
Light energy fuels photosynthetic primary productivity, but its impact on the degradation of algal biomass by complex communities is not well understood. In a recent special Research Topic in Frontiers in Microbiology, we published the results of pairing isotope tracing with NanoSIMS analysis to resolve activity to the single taxon level (using Chip-SIP) to examine the role of light in bacterial processing of algal organic matter. Our experiments used 13C and 15N labeled freeze-thawed diatom cells incubated in laboratory microcosms with a microbial community freshly-collected from the Pacific Ocean, comparing light and dark conditions. To determine the taxon-specific flux of particulate N and C into bacteria, we carried out isotope incorporation analysis by phylogenetic microarray and NanoSIMS (Chip-SIP) to obtain taxon-specific incorporation and combined it with the relative abundance of those taxa from 16S sequencing to calculate total incorporation. We further tested whether taxon-specific isotope incorporation was significantly different according to treatment (light vs. dark or attached vs. free-living). Some taxa, including members of the Flavobacteriaceae and Cryomorphaceae, exhibited increased isotope incorporation in the light, suggesting the use of photoheterotrophic metabolisms. In contrast, some members of Oceanospirillales and Rhodospirillales showed decreased isotope incorporation in the light, suggesting that their heterotrophic metabolism, particularly when occurring on particles, might increase at night or may be inhibited by sunlight. These results show that the transfer between particulate and free-living phases are likely affected by external factors that change with the light regime, such as time of day and season.
[Gomez-Consarnau, L., D.M. Needham, P.K. Weber, J.A. Fuhrman, and X. Mayali, Influence of light on particulate organic matter utilization by attached and free-living marine bacteria. Frontiers in Microbiology, 2019. 10(1204).]
May 2019
Nuccio wins best poster at genomics meeting

On April 5, Erin Nuccio won “Best Overall Poster” at the 2019 Department of Energy Joint Genome Institute’s “Genomics of Energy and Environment Meeting” in San Francisco, CA. Her poster was entitled “Niche differentiation is spatially and temporally regulated in the rhizosphere,” and describes work done at LLNL on the metatranscriptomics of decomposition surrounding the roots. Congratulations Erin!
New Publications
A new collaborative paper between the LLNL team and the Sandia team, co-first authored by Carolyn Fisher and Chris Ward, was published in Algal Research. We investigated the impact of microbiomes on the grazing of Microchloropsis algae by rotifers, and found that specific bacterial taxa were associated with communities that prevented grazing. These protective communities led to the rotifers appearing “sick” and swimming slowly, compared to the controls, where they swam rapidly and consumed algal cells. Adding such microbes could one day be used to pre-treat algal ponds before rotifers colonize them.
Another paper in Algal Research was also just published, first authored by Jeff Kimbrel. In this work, the SFA team investigated the influence of stochasticity on the community assembly dynamics of the algal microbiome, with special focus on the algal phycosphere. In particular, the study found that algal species as well as physical association played significant roles in assembly processes, and that the source community diversity also influenced down-stream patterns of community structure.
Another SFA paper in BMC Bioinformatics on the metabolically versatile Rhodopseudomonas palustris was just published. This amazing organism can switch from 4 different metabolic modes: photoautotrophy, photoheterotrophy, chemoautotrophy, and chemoheterotrophy. Using multi-objective flux analysis modeling, the SFA team determined that R. palustris is light but not carbon limited under anaerobic photoheterotrophic conditions. Of particular interest to the production of biomass, the results of the study indicate that during phototrophic metabolism, optimum allocation of resources and production of ATP are more important than growth.
March 2019
New Publication on how beetle gut microbes make food and fuel

When most people think of wood eating insects, they imagine termites that can destroy a home or business.
However, as part of a Lawrence Livermore National Laboratory bioenergy study, project scientist Jennifer Pett-Ridge and collaborators have learned how the digestive system of a wood-eating beetle serves as a natural mini-reactor for biofuel production.
Led by Javier A. Ceja-Navarro from Lawrence Berkeley National Laboratory (LBL), the team looked at the microorganisms living within the digestive tract of ‘Horned passalus’ beetles (Odontotaenius disjunctus), which live in fallen logs (harvested in Louisiana from fallen pecan tree logs) and are “subsocial”—living in colonies within the wood. These beetles, which are common throughout the Eastern U.S., are responsible for decomposing a huge amount of wood and transforming it into soil organic matter, which contributes to the health and nutrient stock in forest soils. The fact that these beetles can live on food (wood) that is very poor quality—comprised mainly of lignin, cellulose, and hemicellulose with very little nitrogen—is thought to be linked to the capabilities of the bacteria and fungi that reside within their digestive system.
“Understanding how the gut microbiome populations interact to deconstruct lignocellulosic materials to sugars or potential biofuels such as hydrogen and methane could potentially aid in the optimization of industrial cellulosic degradation,” said LLNL biogeochemist Jennifer Pett-Ridge, a co-author on a paper appearing in the journal Nature Microbiology.
The beetle processes large amounts of woody biomass and has developed symbiotic relationships with gut microbes to survive on a low-nutrient diet. The microbial community in the beetle’s gut provides digestive enzymes that degrade complex polysaccharides and lignin of plant cells. Physically, the gut is highly compartmentalized, resulting in a segmentation of microbial populations and the processes for energy extraction from lignocellulose.
To understand the complex connections that make up the O. disjunctus digestive system, the team defined the environment in which these organisms interact, identified the location of key species and their genomes, and confirmed the metabolic pathways contributing to their efficient metabolism of lignocellulosic material.
Breaking down lignin and cellulose to make fuel (sugars etc.) is chemically very difficult, as is fermenting it into a useable reduced product (like acetate, methane or hydrogen). The beetle can’t do this on its own. It needs a diverse group of bacteria and other microbes in its gut to help. The spatial stratification in the beetle’s gut allows for different microenvironments—some are more acidic, some more anaerobic. Each appears to have a community that sequentially degrades the wood substrates and extracts energy, sharing some products with the beetle host.
“We study them because they are a natural model biorefinery,” Pett-Ridge said. “Understanding how evolution has solved the complex lignocellulose to fuel conversation process can help us design better industrial mimics and find novel enzymes or pathways.”
Coarse woody debris makes up a tremendous amount of biomass in forest ecosystems. As a result, several insect groups have evolved to take advantage of this abundant resource, forming specialized wood-feeding guilds. Among the most prominent groups of insects that have specialized in this manner are termites and specific groups of beetles. These groups are considered economically important insects due to the destructive character of some of their members, as examples, subterranean termites, powderpost beetles, bark beetles and longhorn beetles. These insects are able to perform mechanical and enzymatic breakdown of coarse woody biomass that is critically important to the ecology of these ecosystems. This ability to subsist on woody biomass, is in part due to the symbiotic associations that have evolved between these insects and their gut microorganisms.
“These passalid beetle gut communities represent an excellent model system for determining how microbial communities assemble and partition processes along physicochemical gradients to yield an efficient system that converts lignocelluloses into hydrogen, methane, ethanol and other bioenergy-relevant products,” Pett-Ridge said.
Other institutions contributing the research include University of California, Berkeley, Purdue University, Louisiana State University, University of South Carolina, Pacific Northwest National Laboratory and Oregon State University.
The work was funded by the Department of Energy’s Office of Science.
For more information, read the article in The Daily Californian.
[Ceja-Navarro JA, Karaoz U, Bill M, Hao Z, White RA, Arellano A et al (2019). Gut anatomical properties and microbial functional assembly promote lignocellulose deconstruction and colony subsistence of a wood-feeding beetle. Nature Microbiology.]
SFA team member chairs session on photoheterotrophy
Photoautotrophy-dominated environments, such as natural ecosystems like lake, river, and ocean surfaces, as well as engineered ecosystems like photobioreactors, algal turf-scrubbers, and outdoor algal raceways, support entire ecosystems fueled by photosynthesis and dominated by heterotrophic bacterial processes feeding on this recently-fixed carbon. Many heterotrophic bacteria in these ecosystems harbor the genetic machinery to harvest light energy themselves, a process called photoheterotrophy. SFA team member Xavier Mayali recently co-chaired a session at the American Society of Limnology and Oceanography annual meeting in February (with Laura Gomez-Consarnau from the Centro de Investigación Científica y de Educación Superior de Ensenada, Baja California, Mexico) to bring together experts in this growing area of research. Recent discoveries on this topic presented in the session include a newly-discovered type of rhodopsin and quantification of total energy absorbed by photoheterotrophic processes compared to chlorophyll, which further suggest photoheterotrophy is a major global process and may also be important in algal raceway ponds.
February 2019
New Publication on bacterial genome annotation
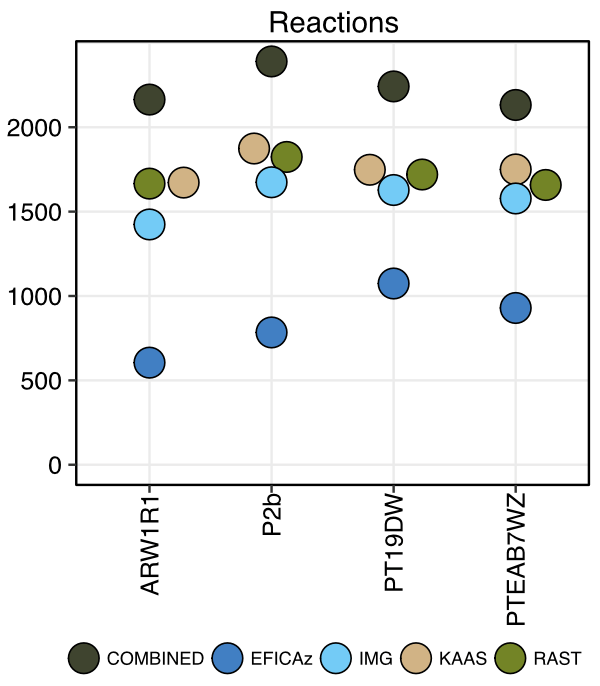
Genome-scale metabolic modeling is a cornerstone of systems biology analysis of microbial organisms and communities, yet these genome-scale modeling efforts are invariably based on incomplete functional annotations. Annotated genomes typically contain 30–50% of genes without functional annotation, severely limiting our knowledge of the “parts lists” that the organisms have at their disposal. In our recent publication, we present results on a comprehensive reannotation of 27 bacterial reference genomes, focusing on enzymes with EC numbers annotated by KEGG, RAST, EFICAz, and the BRENDA enzyme database, and on membrane transport annotations by TransportDB, KEGG, and RAST. Our analysis shows that annotation using multiple tools can result in a drastically larger metabolic network reconstruction, adding on average 40% more EC numbers, 3–8 times more substrate-specific transporters, and 37% more metabolic genes. These results are even more pronounced for pathways outside of the core carbon metabolism, and for bacterial species that are more phylogenetically distant from well-studied model organisms such as E. coli. Our team is currently developing tools for the DOE Systems Biology Knowledgebase to allow users to upload functional annotations from popular third-party annotation tools, compare and merge them, and use them for metabolic modeling.
[Griesemer M, Kimbrel JA, Zhou CE, Navid A, D’haeseleer P, Combining multiple functional annotation tools increases coverage of metabolic annotation, BMC genomics 19 (948), 2018.]
November 2018
Welcoming two new Lawrence Scholar graduate students to LLNL


New Lawrence Scholars Colleen Hui (left) and Katie Harding (right)
SFA team members are hosting two new Lawrence Scholar graduate students.
Colleen Hui is joining us from Sabeeha Merchants group at UC Los Angeles/UC Berkeley. Her research has centered on understanding iron homeostasis in the unicellular green alga Chlamydomonas reinhardtii. At LLNL she will work with NanoSIMS lead Peter Weber on imaging intracellular trace metal metabolism in Chlamydomonas. Specifically, she will be using LLNL’s unique NanoSIMS capability to 1) distinguish the iron storage site in Chlamydomonas cells, 2) analyze the dynamics of intracellular iron movement, and 3) investigate the roles of three candidate vacuole-associated proteins (NRAMP4, CVL1, CVL2) in relation to iron storage and transport in Chlamydomonas. The outcome of this project will provide a better understanding on trace metal utilization in a model alga for sustainable biofuel production.
Katie Harding, a graduate student in Jon Zehr’s group at UC Santa Cruz, is joining the SFA group as a part-time Livermore scholar. Her thesis research is focused on marine nitrogen fixation by unicellular bacteria. At LLNL she will work with Xavier Mayali on using geneFISH (fluorescent in situ hybridization for functional genes) to link microbial functional gene presence and biological activity using NanoSIMS and isotope probing.
SFA team member presents at 2018 International Conference on Microbiome Engineering
SFA member Ali Navid attended the 2018 International Conference of Microbiome Engineering in Boston, MA, in November to present some of his work on computational modeling of microbe-algae and microbe-plant interactions. He presented a talk entitled “Examination of metabolic interactions between phototrophs and their symbiotic microbiome” in the session “From genome-scale to community-scale models.” Learn more about the conference by visiting the 2018 International Conference of Microbiome Engineering website.
October 2018
New Publication on niche partitioning in microbial mats identified using metagenomic analyses
In this study, we examined hypersaline microbial mat communities, which are known for their taxonomic and metabolic diversity and steep environmental gradients. Here, we illustrate how metagenome sequencing can be used to meaningfully assess microbial ecology and genetic partitioning in these complex communities, using binning to reconstruct organisms in silico to assess ecosystem partitioning. We were able to distinguish putative core and accessory genes for the dominant Cyanobacteria in the system, Coleofasciculus chthonoplastes, indicating highest differentiation in nutrient utilization and stress response, suggesting salinity, metals, and light may drive differentiation in this group. We also identified a distinct set of glycoside hydrolases in C.chthonoplastes, building on previous SFA research showing the importance of these polysaccharides in carbon cycling in the mats. The analysis also uncovered evidence of putative phototrophs within the Gemmatimonadetes and Gammaproteobacteria.
[Lee, JZ, Everroad, CR, Karaoz, U, Detweiler, AM, Pett-Ridge, J, Weber PK, Prufert-Bebout, L, Bebout, BM. Metagenomics reveals niche partitioning within the phototrophic zone of a microbial mat. PLoS ONE 13(9): e0202792. 2018]
Collaboration to study sustainable bioenergy agriculture

The LLNL Biofuels SFA is collaborating with Dr. Christine Hawkes (NC State) to improve our understanding of sustainable bioenergy agriculture—studying plant symbionts and belowground processes such as water use efficiency and soil C storage. In a recent experiment, LLNL post-college appointee Max Li worked with the Hawkes lab to harvest Panicum virgatum plants from a study of how foliar fungal endophytes affect plant drought physiology. These endophytes range from antagonistic to beneficial, and are expected to affect the plant’s physiology, biomass, and root metabolites/gene expression.
August 2018
New Publication on carbon fixation enhancement in two species of microalgae by attached microbiomes
NanoSIMS image of a P. tricornutum and heterotrophic bacteria co-culture. The left panel shows the microalgae enriched in freshly fixed carbon, the middle panel shows the varied morphologies of active bacteria comprising the microbiome, and the right panel is a merged view.
We observed mutualistic interactions between heterotrophic bacteria and two species of biofuels-relevant microalgae, Nannochloropsis salina and Phaeodactylum tricornutum, mediated by physical association between individual cells. At the bulk scale, microalgae in these co-cultures exhibited enhanced growth and yield. At the microscale, we used the LLNL NanoSIMS to observe that both species exhibited enhanced carbon fixation in response to the presence of the microbiomes, but there were divergent responses by each species to bacterial attachment. We illustrate how P. tricornutum may be predisposed to interact mutualistically with bacteria via attachment, but N. salina does not share these traits. Attached bacteria benefit from these relationships by receiving more reduced carbon from their algal host compared to free living cells.
[Samo TJ, Kimbrel JA, Nilson DJ, Pett-Ridge J, Weber PK, Mayali X. Attachment between heterotrophic bacteria and microalgae influences symbiotic microscale interactions. Environmental Microbiology 2018, doi: 10.1111/1462‐2920.14357]
Master’s thesis defense

SFA team member Adam Chorazyczewski, a master’s student with Dr. Paul Zimba at Texas A&M successfully defended his master’s thesis entitled “Do phycosphere-associated bacteria affect the growth and lipid accumulation of Phaeodactylum tricornutum.” His SFA-funded research involved profiling and characterizing growth and lipid accumulation in co-cultures of P. tricornutum and 16 separate bacterial species provided by LLNL team members. He identified bacterial isolates with positive effects on both growth and single cell lipid content. SFA algae-bacterial lead Xavier Mayali served as a committee member.
EMSL proposal awarded
SFA team members led by Ty Samo were awarded an EMSL grant entitled “Nano- to microscale characterization of metabolic cooperation facilitated by physical associations between phototrophic microalgae and heterotrophic bacterial symbionts.” Co-Investigators include R. Stuart, X. Mayali, C. Ward, P. Weber, T. Northen (LBL), and C. Buie (MIT).
June 2018
Algae Outreach Event at LLNL
SFA team members Carolyn Fisher (Sandia), Xavier Mayali (LLNL), and Chris Ward (LLNL) helped host a hands-on work station, called “Algae Invader Investigation,” on algae and algal predators. Students learned about bioenergy research on algae and the current cultivation obstacles, such as algal predators. They used both microscopes and household items such as beans and pasta to demonstrate how algae are studied in the lab. According to the Sandia summer intern Franny Carcellar: “I had one girl tell me after looking through the microscope that now she wants to be a scientist. If we just inspire one kid, then it’s all worth it!”
For more information on the event, see the article titled “STEM Day at the Lab gives underserved students a taste of the wonders of science” on the LLNL website.


May 2018
New publication on multimodal LA-ICP-MS and nanoSIMS imaging
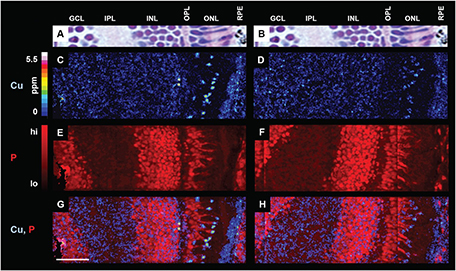
We are developing high sensitivity, high spatial resolution methods to image essential metals in biological systems. Here, we collaborated with the C. Chang Lab at UC Berkeley to standardize our NanoSIMS copper measurements and apply the method to analysis of copper metabolism in zebrafish. Using this method, we were able to demonstrate that the fish metal distribution system prioritizes delivering copper to the eye, despite a severe copper deficit caused by a genetic mutation which mimics a human copper dysregulation disorder, Menkes disease.
[Ackerman CM, Weber PK, Xiao T, Thai B, Kuo TJ, Zhang E, Pett-Ridge J, Chang CJ. Multimodal LA-ICP-MS and nanoSIMS imaging enables copper mapping within photoreceptor megamitochondria in a zebrafish model of Menkes disease. Metallomics 2018 10(3):474-85.]
Greenhouse at LLNL now open!

Our new greenhouse facility is now operational. It is a new IGC Arch Series 6500 greenhouse (1800 sf) with full temperature and light controls. It will eventually house an array of Coy isotope labeling chambers for plant 13CO2 isotope labeling and our Picarro portable Cavity Ring-Down Spectroscopy (CRDS) analyzer. We are currently setting up a series of plant-AMF inoculations.
Invited seminar on computational systems biology research

SFA team member Ali Navid recently gave an invited seminar as part of the LLNL’s and nearby Las Positas College student science and engineering seminar series. His talk, given to a large audience of community college science students and faculty, was titled “Computational Systems Biology: Simulating life from microbes to humans.” He discussed the state of computational systems biology in general and particularly at LLNL and presented examples of different types of modeling (e.g., constraint-based genome-scale, dynamic pharmacology, and 3D microbial biophysics models) that span multiple time and spatial scales from his SFA and biosecurity-related research.
April 2018
New Publication: Quantitative isotope incorporation reveals substrate partitioning in a coastal microbial community
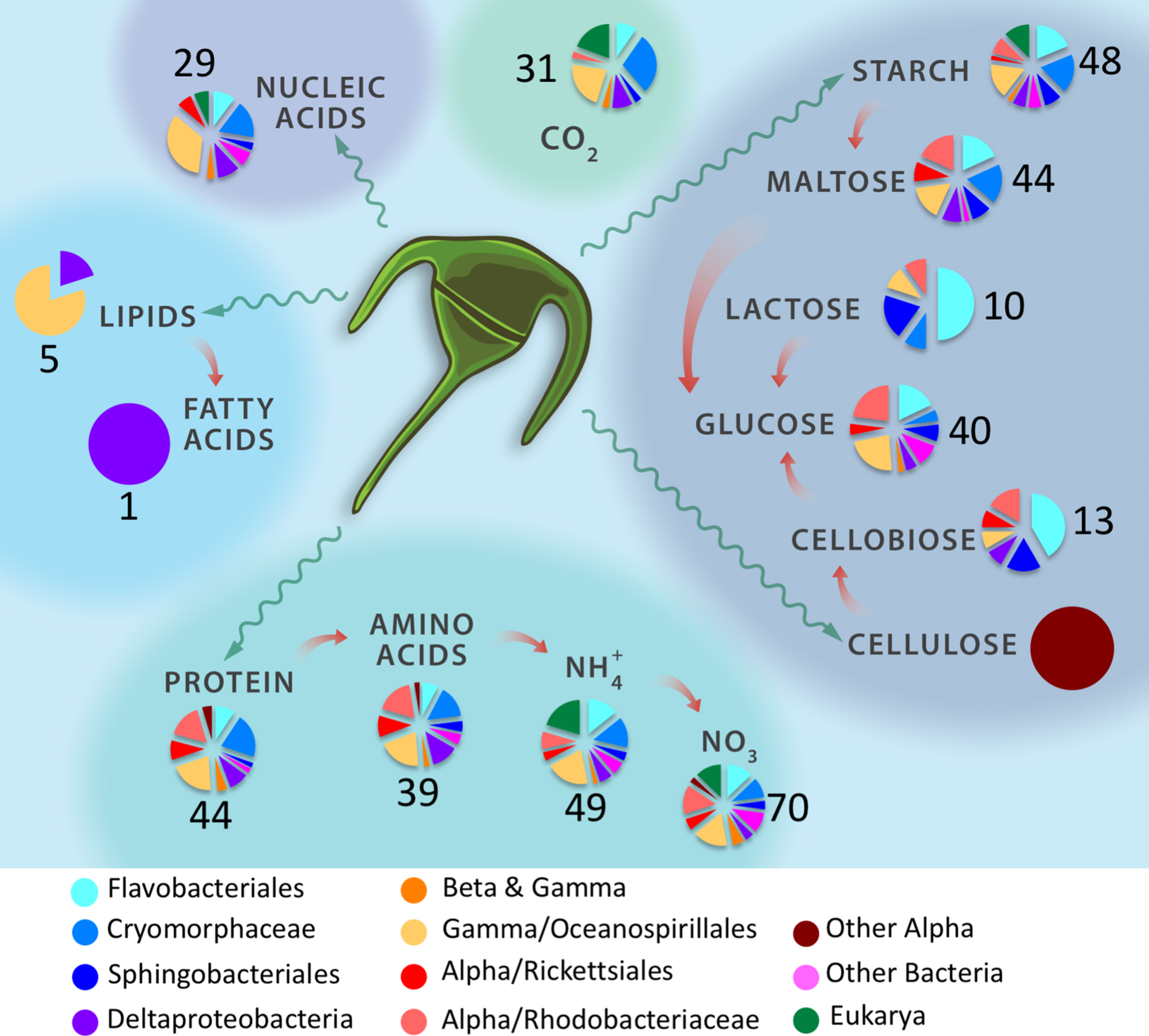
SFA team members Xavier Mayali and Peter Weber have a new publication, supported by the Biofuels SFA, on the use of the Chip-SIP method (NanoSIMS and microarrays) to quantify the taxon-specific incorporation of algal-derived organic components by bacteria and eukaryotes. They carried out simultaneous incubations with 14 different stable isotope labeled substrates to examine phylogenetic signal of resource utilization and mixotrophy. The unique aspect of examining such a high number of substrates enabled them to identify substrate partitioning, and the data showed that two thirds of the taxa exhibited unique incorporation patterns, with strategies ranging from generalists to specialists.
[X. Mayali and P.K. Weber, Quantitative isotope incorporation reveals substrate partitioning in a coastal microbial community, FEMS Micro Ecol 94 (5), fiy047 (2018), doi:10.1093/femsec/fiy047]
March 2018
SFA team members attend annual JGI user meeting

Five SFA team members presented at the 13th annual Department of Energy Joint Genome Institute (JGI) “Genomics of Energy and Environment” meeting in San Francisco, CA. Three posters were presented: Chris Ward, “Towards an integrative understanding of chytrid parasitism and its drivers in mass algal culture,” Jeff Kimbrel, “Combining multiple functional annotation tools increases completeness of metabolic annotation,” and Xavier Mayali, “Nanoscale Stable Isotope Tracing to Investigate Interactions between Bacteria and Biofuel-producing Algae.” Mayali’s poster won the “Outside the Box Poster Award.” The LLNL team also presented two invited talks: “Exploring Microbial Ecology with Isotopes and Imaging” by Jennifer Pett-Ridge and “Exploring Metabolite Production from Tryptophan Precursors in Two Algal Associated Bacteria, Algoriphagus sp. ARW1R1 and Marinobacter sp. 19DW” by Ty Samo.
Visit the JGI website for more information.
LLNL’s Biofuels SFA is hiring!
We have a position for a postdoc to examine host-microbe metabolic interactions and exchange in microalgae and perennial grasses using metabolomics, stable isotope probing, and proteogenomic approaches. For more information, see position 103155 at https://jobs.llnl.gov. If you have questions, feel free to contact Michael Thelen at thelen1 [at] llnl.gov (thelen1[at]llnl[dot]gov).
February 2018
New publication: High initial sputter rate found for Vaccinia virions using isotopic labeling, NanoSIMS, and AFM
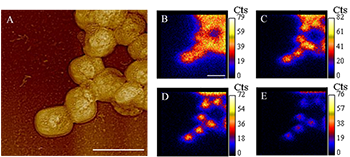
Correlated AFM and SIMS images of Vaccinia virions showing colocalization in the two instruments and general nature of virion surface erosion during sputtering. The AFM image (A) is a contrast-enhanced height image that corresponds to the region in the SIMS images (B–E). The sequence of SIMS images detail the distribution of the 12C14N- species counts during each scan. SIMS image (B), (C), (D), and (E) correspond to scan number 1, 15, 25, and 39, respectively. The decrease in species counts as a function of scan number is a result of lateral erosion caused by sputtering. Scale bar: 500 nm.
New research supported by the LLNL Biofuels Scientific Focus Area (SFA) shows the promise and challenge of studying the role of viruses in microbial systems. Viruses are known to be ubiquitous and infect all forms of life, including microbes. As such, they are thought to have major roles in carbon and nutrient cycling, but to date, these rates have been unexplored in most systems. With this research, SFA researchers Peter Weber and Ben Stewart and colleagues explored the potential to use high-spatial resolution secondary ion mass spectrometry (SIMS) with a NanoSIMS 50 to characterize nutrient transfer from host to virus using isotopically labeled DNA as a tracer. Their work showed the expected transfer of isotopic label, which is promising for future application of SIMS to virus ecology, but they also found that at the start of SIMS analysis, these tiny structures eroded 100 times faster than previously expected. For the relatively large Vaccinia virus in this study, this was not a major problem, but for bacterial phage, which are as small as 20 nm in diameter, new methods may be necessary to ensure quality measurements of nutrient uptake and cycling.
[S. Gates, R.C. Condit, N. Moussatche, B.J. Stewart, A.J. Malkin, and P.K. Weber, High Initial Sputter Rate Found for Vaccinia Virions Using Isotopic Labeling, NanoSIMS, and AFM, Anal. Chem. 90 (3), 1613 (2018), doi: 10.1021/acs.analchem.7b02786]
SFA team members present at 2018 Ocean Sciences meeting
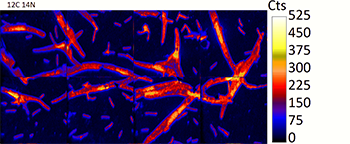
An ion image generated from the NanoSIMS illustrates the high spatial resolution visualization of algal and bacterial biomass, as determined from 12C 14N ion masses. Using these images, exchanges of C and N between algae and bacteria as a function of their spatial interactions can be measured.
SFA members Xavier Mayali and Ty Samo attended the 2018 Ocean Sciences meeting in Portland, Oregon in February to present some of their work on algal-bacterial interactions using stable isotope probing and NanoSIMS. Ty presented a talk entitled “Stable Isotope Probing and NanoSIMS Reveals Effects of Physical Association on Mutualisms Between Individual Bacterial Cells and Two Species of Phytoplankton” in the session “Bridging Microbial, Stable Isotope, and Micronutrient Approaches to Marine Carbon and Nitrogen Recycling.” Xavier presented a talk entitled “Investigating C transfer between diatoms and phycosphere-associated bacteria with stable isotopes and NanoSIMS” in the session “Phytoplankton-Bacteria Interactions: From Microscales to Ocean Scales.”
Learn more about the conference on the 2018 Ocean Sciences Meeting site.
What Can Algae Do For Us?
In this video, SFA scientists help explain what algae are and what they can do for us.